A data structure shared by databases supporting multiple taxonomic views developed for different taxa: a comparative analysis
Nozomi Ytow (Institute of Biological Sciences, University of Tsukuba, Japan)
David McL. Roberts (Department of Zoological, The Natural History Museum, UK)
David R. Morse (Computing Department, Faculty of Mathematics and Computing, The Open University, UK)
Abstract
Interconnecting databases of names, taxa or collections requires that they interoperate. This includes providing support for multiple taxonomic views because it is inconceivable that all databases should use the same, single taxonomic view of the data. A wide coverage of data, a diversity of intended audiences or even a long period of development can result in multiple taxonomic views being provided in a single database. Therefore, the ability to support multiple taxonomic views is a core requirement for global-scale interoperability of databases.
Several data models have been developed and proposed independently in recent years. These models include FGDC (ITIS, 2000), Nomencurator (Ytow et al., 2001), Pisces-II (Pyle, 2000) and Prometheus (Pullan et al., 2000). All of them support multiple taxonomic views, which demonstrates an increasing recognition of the necessity to provide such a facility in order to assist taxonomic work. Each of these models inherits some of the customs and practices from the background taxa for which the model was developed. This suggests that the models may be lacking in their ability to support multiple taxonomic views of a global-scale federation of databases. Finally, each of the models also contains elements of the potential taxon concept (Berendsohn, 1995 and 1997).
Characteristics of models
Compared all data structures support multiple taxonomies. Adding that, they have following characteristics:
- FGDC
- Intending to be general coverage model, but rather botanical data model
- Ecological application is also in scope
- Distinction between usage of a name and assertion on a name
-
IOPI
- Botanical database
- Designed to allow data exchange between systems
- Distinction of scientific name and potential taxon
- Status assignment to a potential taxon can be multiple
- Rich in literature data structures
-
Nomencurator
- General coverage model for taxonomic research
- Literature based, name usage tracing with annotations
- Taxonomic data is stored as hierarchy implicitly
- Focusing on fragmented data compilation
- Scalability and saving maintenance effort
-
Pisces-II
- Developed for own research in ichthyology
- Rich in specimen handling structures
-
Prometheus
- Botanical database
- Separation of Nomenclatural Taxon and Circumscribed Taxon by focusing how a taxon concept (and hence a name) is created
- List of specimen or subordinate taxa as testable circumscription
- Automatic name calculation
 Entity or class name are shown in Helvetica. Attributes of class or entity are shown by bold Times. Metastructures are shown in oblique Times.
A colour encodings is also used: green for name related structures, blue for taxonomy, orange for assertion on taxa, brown for instances, purple for publications and authors and red for unsupported. Publications and authors are encoded by the same colour because a publication is a "time-stamped author" (Richard L. Pyle, personal comm.) in essence.
Entity or class name are shown in Helvetica. Attributes of class or entity are shown by bold Times. Metastructures are shown in oblique Times.
A colour encodings is also used: green for name related structures, blue for taxonomy, orange for assertion on taxa, brown for instances, purple for publications and authors and red for unsupported. Publications and authors are encoded by the same colour because a publication is a "time-stamped author" (Richard L. Pyle, personal comm.) in essence.
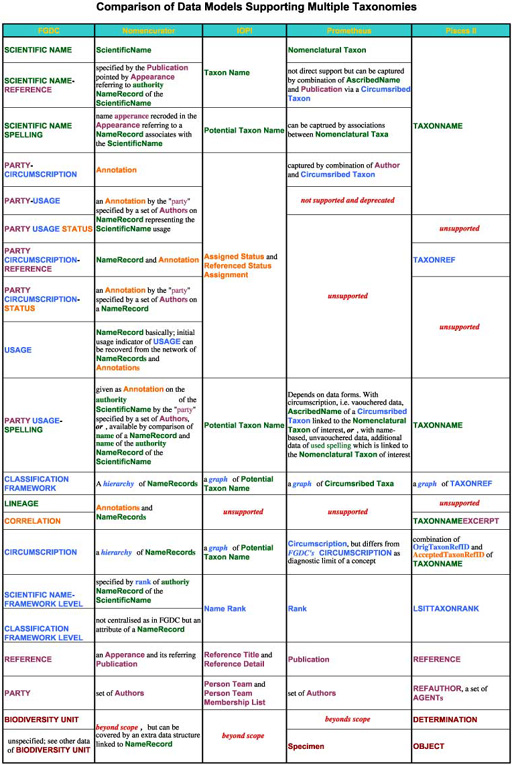
Data coverage varies model by model. Some data structures in a model are covered by relationships of data structures, i.e. meta structure, in other models. Even models with smaller number of structures can have similar coverage. Smaller number of mandate data structure covering wider range of information makes data exchange easier, while larger superset of databases can result in ambiguous assignment of attributes to data structures. All models supports data structure capturing name, taxon concept, publication reference and author under different terminologies. They may not identical but substantially overlap with "equivalents" in other models. Relationships between name and/or taxon concept are also supported by all models somewhat. These data structures composes a candidate of common data model sharable between databases. Terminologies used by data models for these core data structures are summarised in a table.
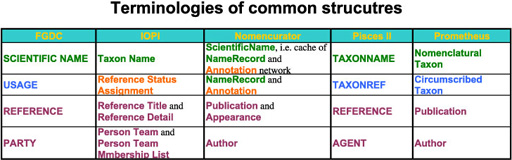
Only differences in terminology?
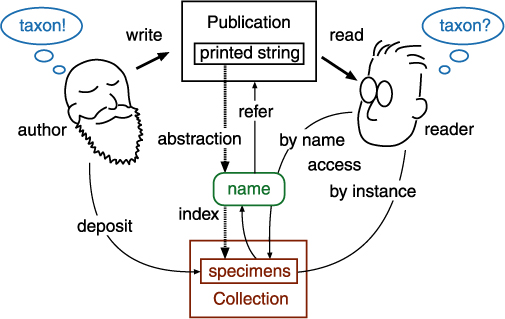
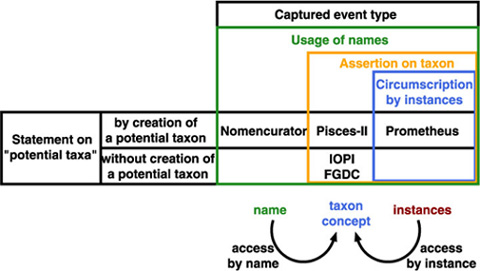
Taxonomic database is an extension of communication between "users" including taxonomists. It captures a summary of a taxonomic work just like taxonomic works appeared in traditional publications. A communication model amongst taxonomists shows that there is no direct link between the taxon concepts of the author and the reader (user). There are two indirect ways to access to a taxon concept, by name or by instance. Circumscription sensu Prometheus captures access by instance, while Nomencurator captures access by name as potential taxon sensu Berendsohn (1995), or, a usage of the name.
 Assertion on taxon concept is another way to access to a taxon concept. While it defines relationship between taxon concepts rather than taxon concept itself, it provides an access key to taxon concept as a result of exclusive nature of classification. We have, therefore, name, taxon concept and instances as target of actions on them, i.e. usage, assertion and circumstription. They compose a nested structure as combination where cercumscription by instances defines taxon concept from inside (lower level) ostensively, while name gives outer circumference of taxon concept.
Assertion on taxon concept is another way to access to a taxon concept. While it defines relationship between taxon concepts rather than taxon concept itself, it provides an access key to taxon concept as a result of exclusive nature of classification. We have, therefore, name, taxon concept and instances as target of actions on them, i.e. usage, assertion and circumstription. They compose a nested structure as combination where cercumscription by instances defines taxon concept from inside (lower level) ostensively, while name gives outer circumference of taxon concept.
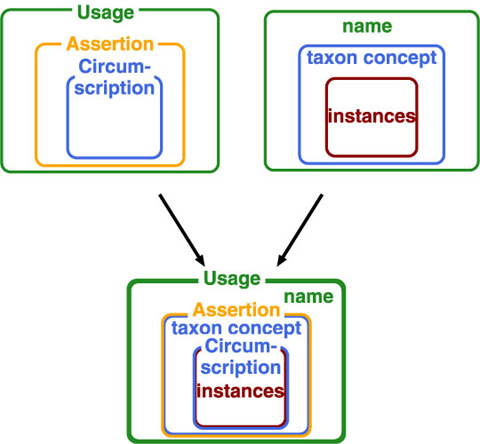
Adding to these action-types, two styles are distinguished: whether action accompanies creation of a potential taxon.
Therefore, models can be placed into matrix of three action types by tow styles. Since a potential taxon is a pair of name and publication (or its equivalent), circumscription and name usage always accompany potential taxon. Although assertion on taxon is also expected to accompany potential taxon creation in that sense, some models enable to give a status to a potential taxon without linkage to another potential taxon considered appropriate by the assigner. Note that circumscription, assertion and usage are nested; therefore, a data model capturing name usage also captures both assertion and circumscription.
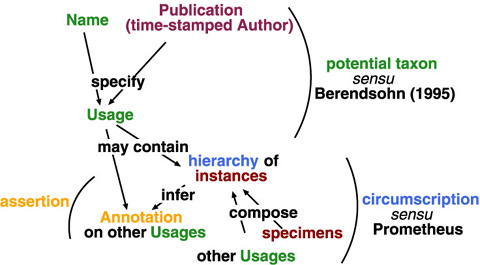
 Since circumscription is a subset of assertion, and assertion is a subset of name usage, sharable common data elements would be organised by usage, with optional circumscription and assertion. A name and a publication (or, time-stamped author) specifies a name usage in sense of potential taxon (Berendsohn, 1995). The name usage may have assertion and circumscription as its contents; indeed, circumscription by specimen requires additional data structure. A circumscription always infers an annotation on existing potential taxa (usages) because taxonomy works as differentiation to already known classifications. Annotations between name usages allow to capture multiple taxonomic views even if circumscriptive detail is unavailable in the given publication.
Since circumscription is a subset of assertion, and assertion is a subset of name usage, sharable common data elements would be organised by usage, with optional circumscription and assertion. A name and a publication (or, time-stamped author) specifies a name usage in sense of potential taxon (Berendsohn, 1995). The name usage may have assertion and circumscription as its contents; indeed, circumscription by specimen requires additional data structure. A circumscription always infers an annotation on existing potential taxa (usages) because taxonomy works as differentiation to already known classifications. Annotations between name usages allow to capture multiple taxonomic views even if circumscriptive detail is unavailable in the given publication.
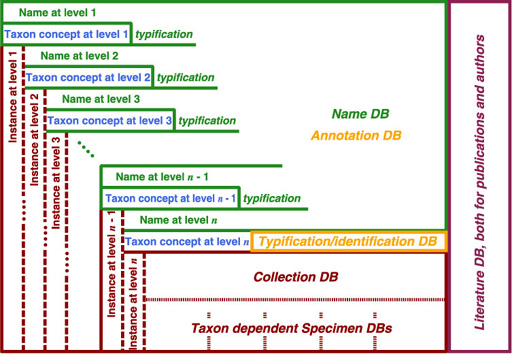
 For names of rank higher than species have names as their instances, instead of specimen for species or lower. Therefore, name usage also covers "access by instance" for higher ranks. It can be captured by a database architecture shown below. Six databases are logically distinguished, but unnecessary to be implemented as separated databases. It is also possible to implement them as separate tables in single RDBMS. Detail of physical implementation depends on volume of data, coverage of taxa and performance. Note that taxon concept at each level is encapsulated by name for higher level or by identification at entity level. Database covering taxon information such as description must be covered separately. The entity level is unnecessary to be species or lower; if you intend to examine family based on specimen instead of genus, the entity level must be shifted to family. It implies that this architecture also works under PhyloCode style taxonomic view.
For names of rank higher than species have names as their instances, instead of specimen for species or lower. Therefore, name usage also covers "access by instance" for higher ranks. It can be captured by a database architecture shown below. Six databases are logically distinguished, but unnecessary to be implemented as separated databases. It is also possible to implement them as separate tables in single RDBMS. Detail of physical implementation depends on volume of data, coverage of taxa and performance. Note that taxon concept at each level is encapsulated by name for higher level or by identification at entity level. Database covering taxon information such as description must be covered separately. The entity level is unnecessary to be species or lower; if you intend to examine family based on specimen instead of genus, the entity level must be shifted to family. It implies that this architecture also works under PhyloCode style taxonomic view.
Acknowledgement
We are grateful to Richard L. Pyle and Mark R. Pullan for detailed explanation of their data structures.
References
-
Potential taxon concept:
Berendsohn WG (1995)
The Concept of Potential Taxa in Databases.
Taxon 44: 207-212.
-
FGDC:
Steve Taswell and Robert Peet(2000)
Biological Nomenclature/Taxonomy Meeting Summary
available from
http://biology.usgs.gov/fgdc.bio/FGDC_A.DOC
Project summary of
Biological Nomenclature and Taxonomy Data Standard is
available from http://www.fgdc.gov/standards/status/sub5_8.html/
-
IOPI:
Berendsohn WG (1997)
A taxonomic information model for botanical
databases: the IOPI Model.
Taxon 46: 283-309.
-
Nomencurator:
Ytow, N, Morse DR, Roberts DM. (2001)
Nomencurator: a nomenclatural history model to handle
multiple taxonomic views.
Biological Journal of Linnean Society
73(1):81-98.
reprint is available from
http://www.idealibrary.com/links/doi/10.1006/bijl.2001.0527/pdf
http://www.nomencurator.org/
-
Pisces-II:
Richard L. Pyle (2000) ASIH meeting
-
Prometheus:
Pullan MR, Watson MF, Kennedy JB, Raguenaud C, Hyam R. (2000)
The Prometheus Taxonomic Model: a practical approach to representing
multiple classifications.
Taxon 49: 55-75.
http://www.prometheusdb.org/







